How can people make good decisions based on limited, noisy information?
FFTrees
An R package to create, visualize, and evaluate fast-and-frugal decision trees.......Or, how I learned to stop worrying and love heuristics.
Nathaniel Phillips, Economic Psychology, University of Basel, Switzerland
SPUDM 2017, Haifa, Israel
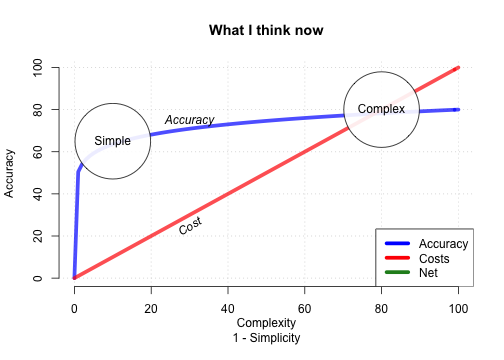
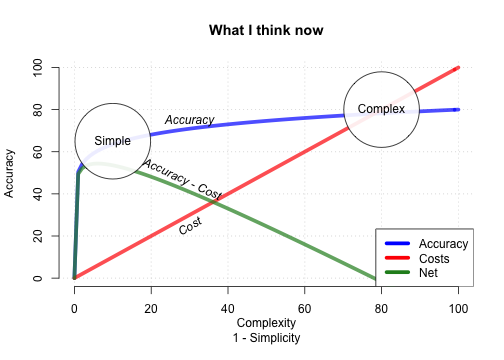
Limited Time. Limited information.

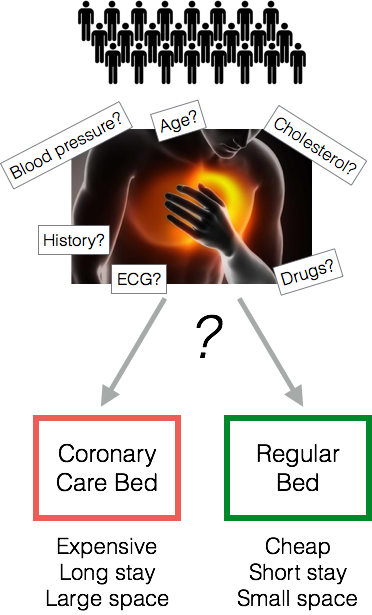
Heart Attack Diagnosis
- How do doctors make decisions? Experience. Intuition. Clinical judgment

- In a Michigan hospital, doctors sent 90% of patients to the ICU, although only 25% were actually having a heart attack.
Emergency Room Solution: a fast-and-frugal tree (FFT)
- A fast-and-frugal decision tree (FFT) developed by Green & Mehr (1997).
- Tree cut the false-alarm rate in half
- Tree is transparent, easy to modify, and accepted by physicians (unlike regression).
What is a fast-and-frugal decision tree (FFT)?
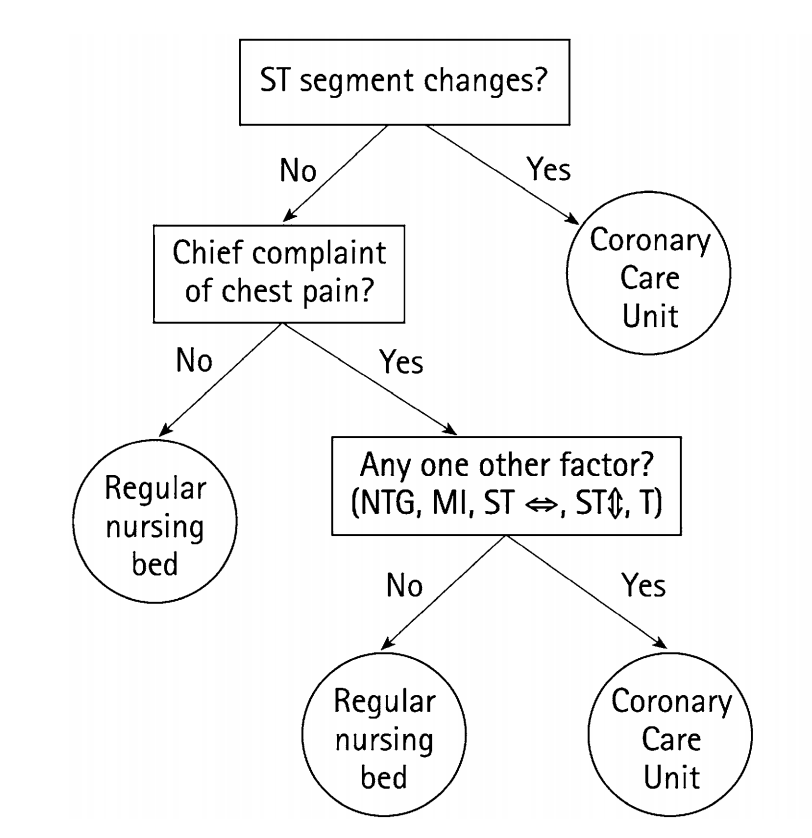
Green & Mehr (1997) "What alters physicians' decisions to admit to the coronary care unit?"
Fast-and-Frugal Decision Tree (FFT)
- An FFT is a decision tree with exactly two branches from each node, where one, or both, of the branches are exit branches (Martignon et al., 2008)
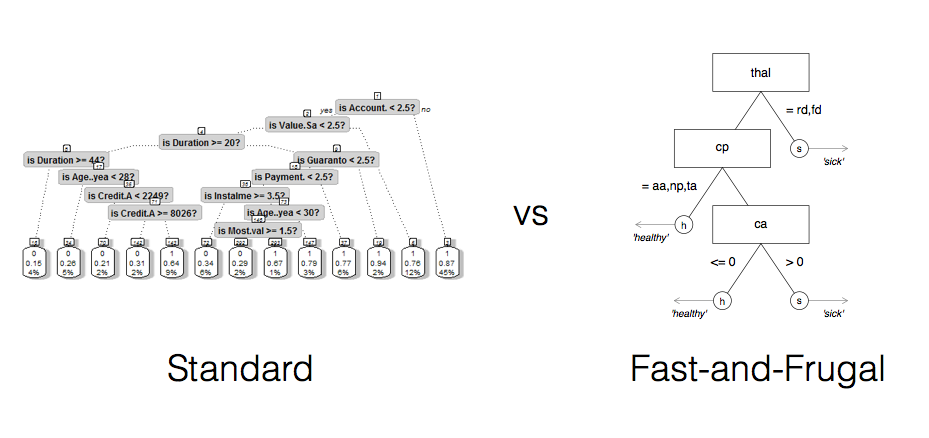
Examples of FFTs
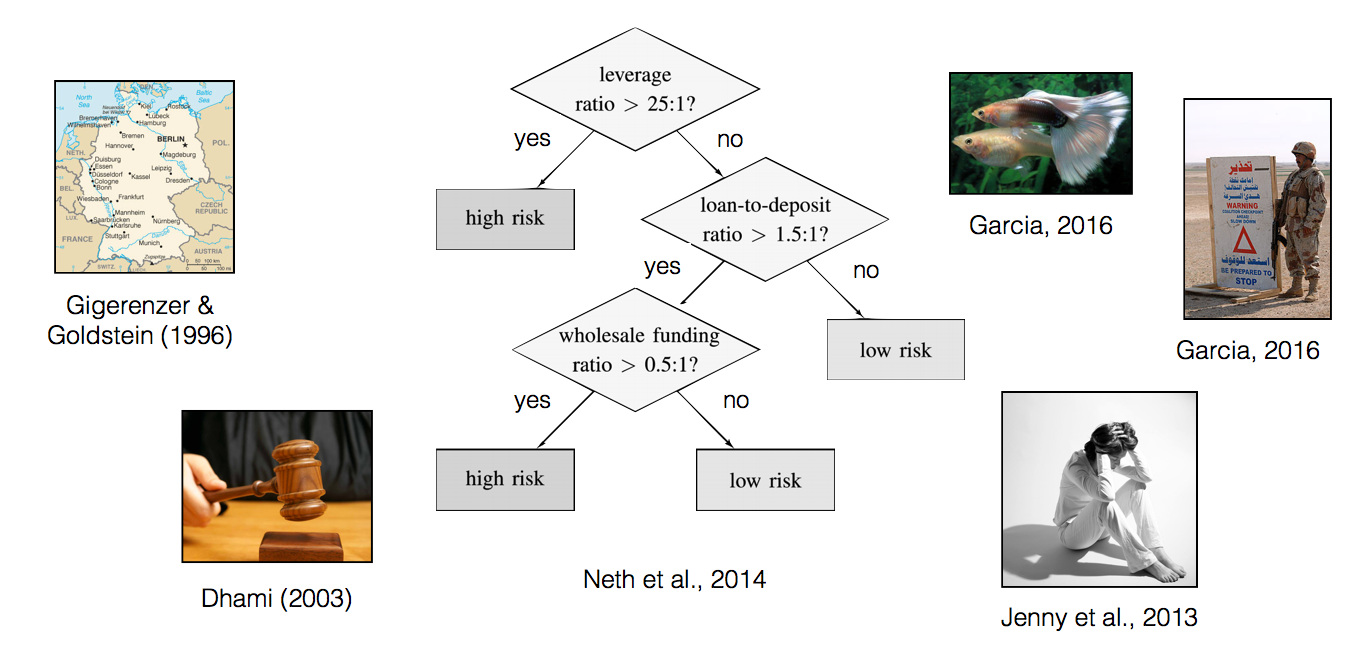
But how can I create FFTs?
- Need an algorithm (and preferably software)
| Algorithms | Software | |
|---|---|---|
| Standard Decision Trees | CART, CHAID | SPSS, Excel, R, Matlab, ... |
| Fast-and-Frugal Trees (FFTs) | Max, Zig-zag (Martignon et al., 2003; 2008) | ? |
Missing: An easy to use toolbox that creates and visualizes FFTs based on data.
Answer: FFTrees

If you don't like things for free (R), you can pay IBM SPSS $680 / year to make standard decision trees.

Martignon, Katsikopoulos & Woike (2008)
FFTrees
- A package to create, visualize, and evaluate fast-and-frugal decision trees.
- Minimal programming, extensive examples and guides, informative visualizations.
# 3 Steps to getting started
install.packages("FFTrees") # Install
library("FFTrees") # Load
FFTrees_guide() # Open guide
0
/ \
F 0
/ \
F T
FFTrees v1.3.3
Tutorial and Documentation
Example: Heart Disease
| age | sex | cp | trestbps | chol | fbs | restecg | thalach | exang | oldpeak | slope | ca | thal | diagnosis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 63 | 1 | ta | 145 | 233 | 1 | hypertrophy | 150 | 0 | 2.3 | down | 0 | fd | 0 |
| 67 | 1 | a | 160 | 286 | 0 | hypertrophy | 108 | 1 | 1.5 | flat | 3 | normal | 1 |
| 67 | 1 | a | 120 | 229 | 0 | hypertrophy | 129 | 1 | 2.6 | flat | 2 | rd | 1 |
| 37 | 1 | np | 130 | 250 | 0 | normal | 187 | 0 | 3.5 | down | 0 | normal | 0 |
| 41 | 0 | aa | 130 | 204 | 0 | hypertrophy | 172 | 0 | 1.4 | up | 0 | normal | 0 |
| 56 | 1 | aa | 120 | 236 | 0 | normal | 178 | 0 | 0.8 | up | 0 | normal | 0 |
Goal: Predict binary diagnosis classification
The FFTrees() function
# Step 1: Install and load FFTrees (v.1.3.2)
install.packages("FFTrees")
library("FFTrees")
# Step 2: Create FFTs
heart.fft <- FFTrees(formula = diagnosis ~., # Formula
data = heart.train, # Training data
data.test = heart.test, # Test data
main = "Heart Disease", # Optional labels
decision.labels = c("Low-Risk", "High-Risk"))
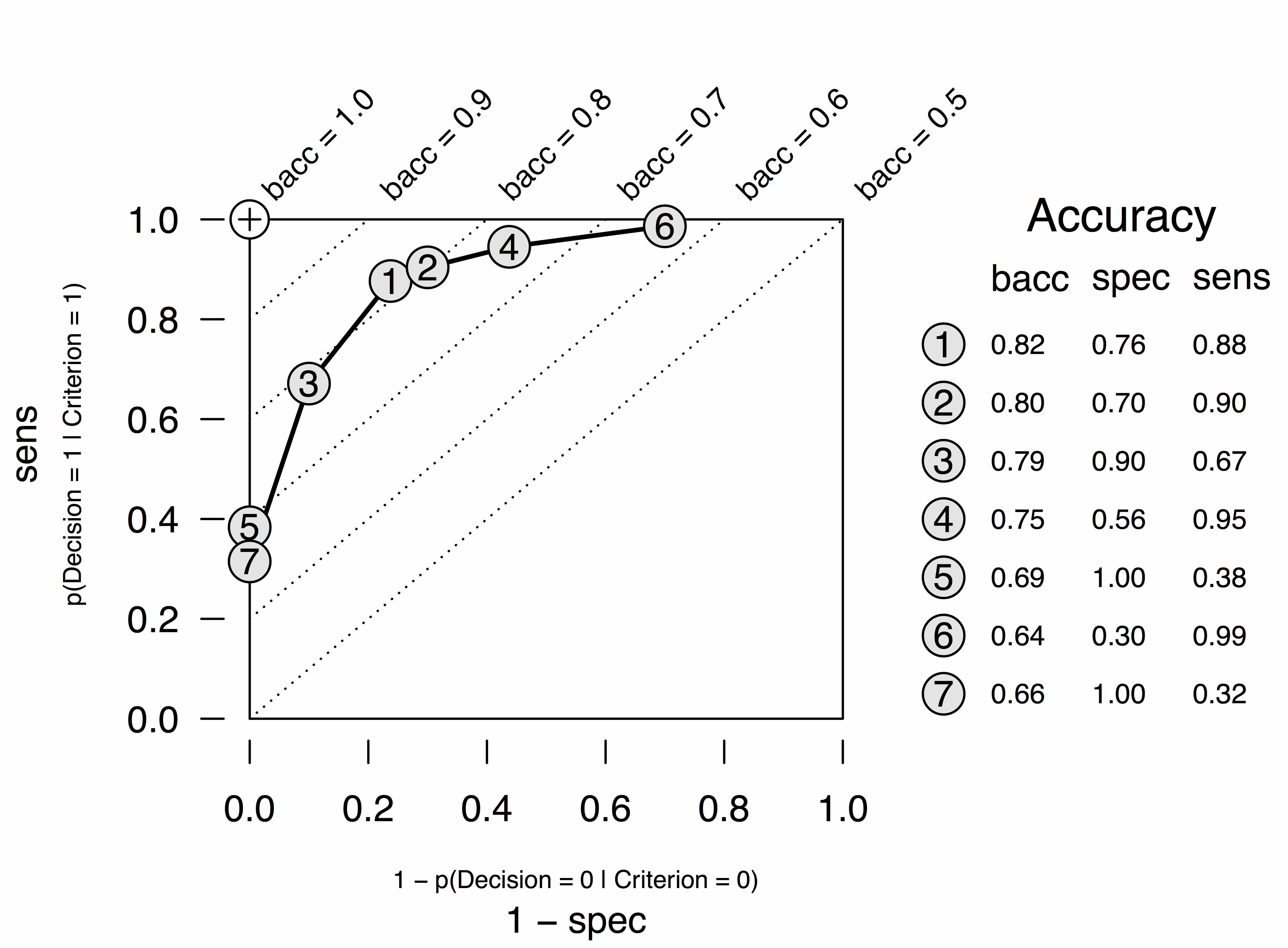
Print an FFTrees object
heart.fft
Heart Disease
7 FFTs predicting diagnosis (Low-Risk v High-Risk)
FFT #1 uses 3 cues: {thal,cp,ca}
train test
cases :n 150.00 153.00
speed :mcu 1.74 1.73
frugality :pci 0.88 0.88
accuracy :acc 0.80 0.82
weighted :wacc 0.80 0.82
sensitivity :sens 0.82 0.88
specificity :spec 0.79 0.76
pars: algorithm = 'ifan', goal = 'wacc', goal.chase = 'bacc', sens.w = 0.5
Print a tree "in words"
inwords(heart.fft)
[1] "If thal = {rd,fd}, predict High-Risk"
[2] "If cp != {a}, predict Low-Risk"
[3] "If ca <= 0, predict Low-Risk, otherwise, if ca > 0, predict High-Risk"
| cue | definition | Possible values |
|---|---|---|
| thal: thallium scintigraphy | How well blood flows to the heart | normal (n)fixed defect (fd), or reversible defect (rd) |
| cp: chest pain type | Type of chest pain | typical angina (ta), atypical angina (aa), non-anginal pain (np), or asymptomatic (a). |
| ca: | number of major vessels colored by flourosopy | 0, 1, 2, 3 |
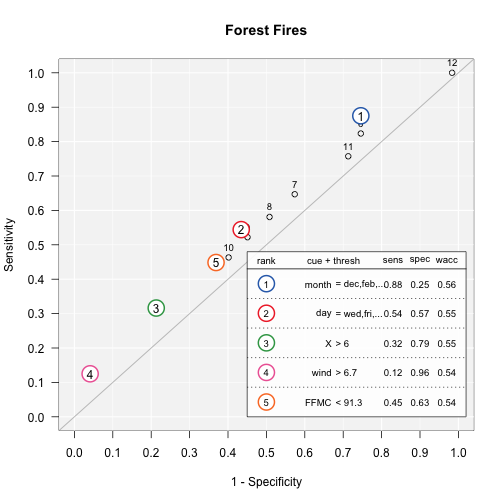
plot(heart.fft, stats = FALSE, data = "test")
## Error in plot(heart.fft, stats = FALSE, data = "test"): object 'heart.fft' not found
plot(heart.fft, data = "test") # Training data
## Error in plot(heart.fft, data = "test"): object 'heart.fft' not found
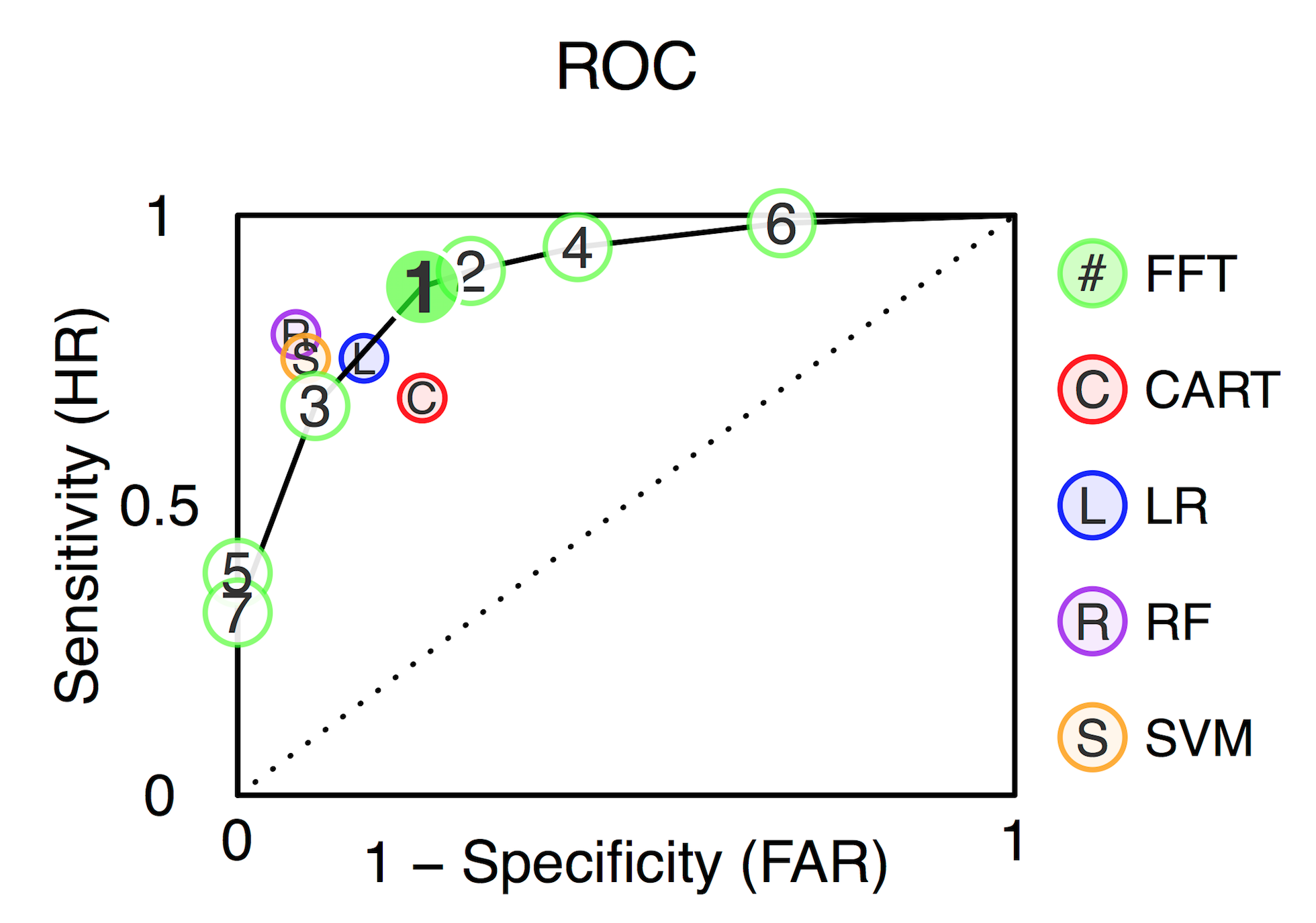
plot(heart.fft, data = "test", tree = 6) # Testing data, tree 6
## Error in plot(heart.fft, data = "test", tree = 6): object 'heart.fft' not found
plot(heart.fft, data = "test", tree = 7) # Testing data, tree 7
## Error in plot(heart.fft, data = "test", tree = 7): object 'heart.fft' not found
How well can simple FFTs compete with classical rational models and cutting-edge machine learning algorithms?
Prediction Simulation
- 10 datasets from the UCI Machine Learning Database.
- 50% Training, 50% Testing
- FFTrees vs rpart, regression, random forests...
- Criterion: Balanced accuracy
mean(sensitivity, specificity)

## Error in plot(mushrooms.fft): object 'mushrooms.fft' not found
## Growing FFTs with ifan
## Fitting non-FFTrees algorithms for comparison (you can turn this off with comp = FALSE) ...
## Growing FFTs with ifan
## Fitting non-FFTrees algorithms for comparison (you can turn this off with comp = FALSE) ...
Speed and frugality
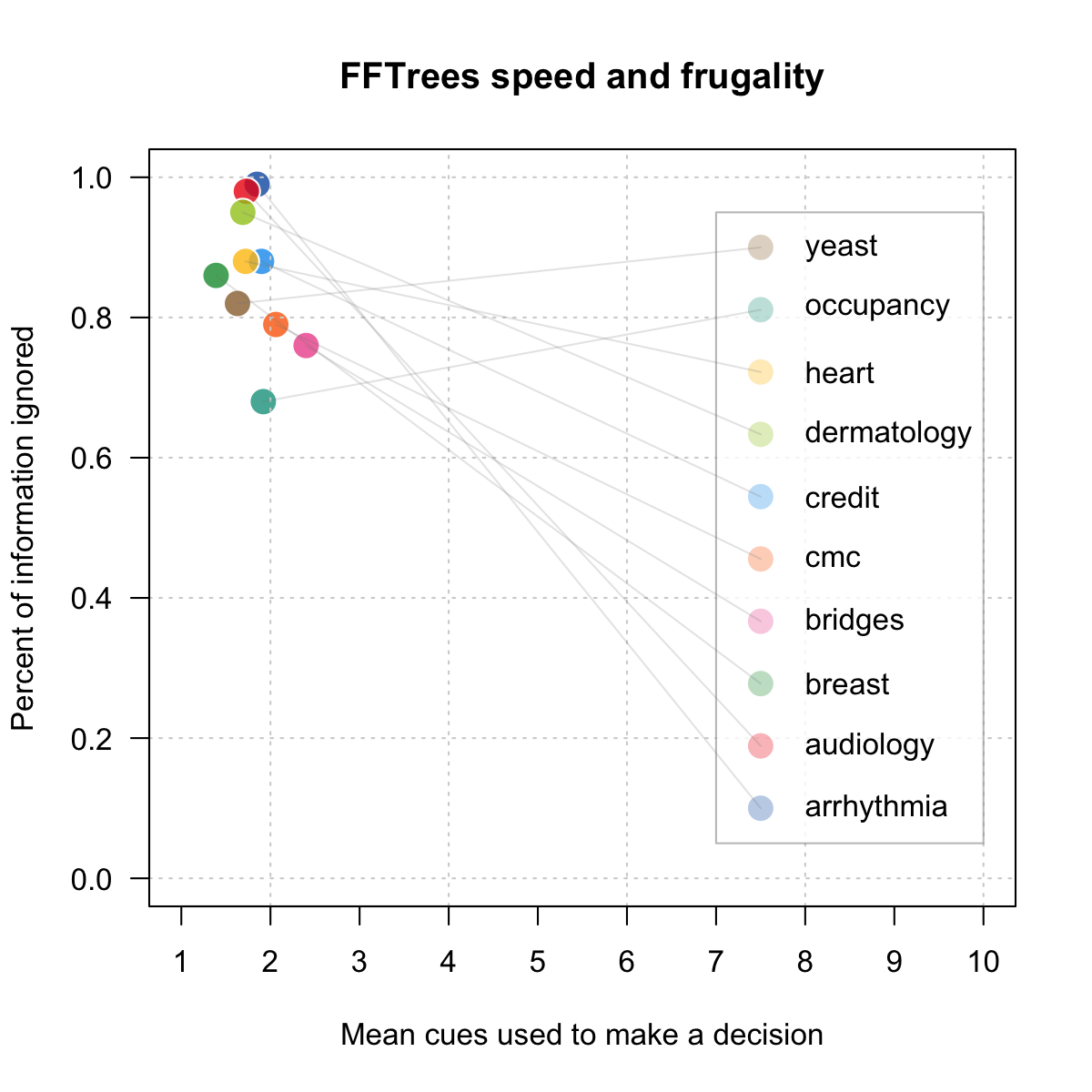
Speed and frugality
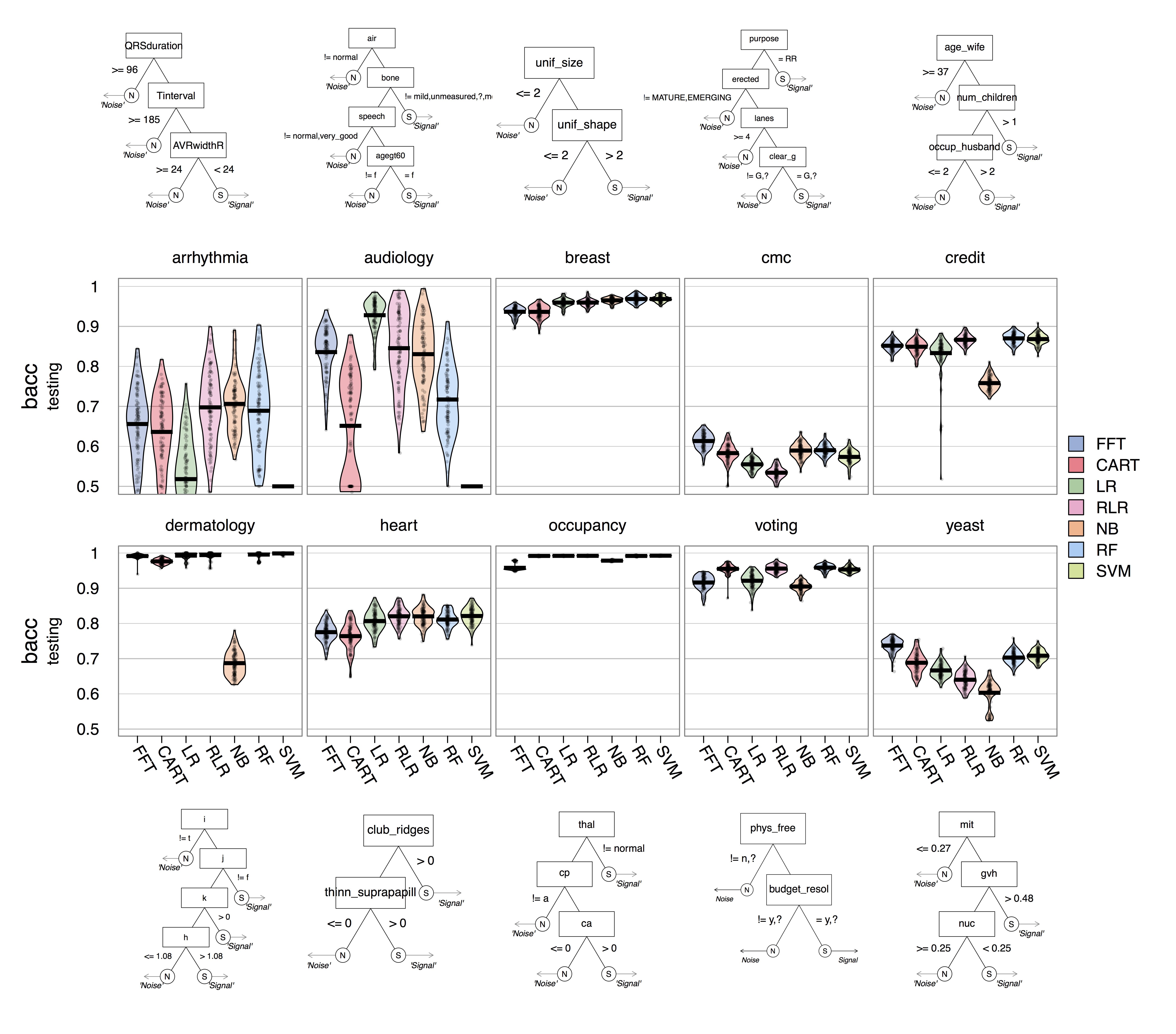
Fitting Accuracy
## Error in subset(FFTrees.mlr.df, ((algorithm == "SVM" & task.id %in% c("arrhythmia", : object 'FFTrees.mlr.df' not found
## Error in int_abline(a = a, b = b, h = h, v = v, untf = untf, ...): plot.new has not been called yet
Prediction Accuracy
## Error in subset(FFTrees.mlr.df, ((algorithm == "SVM" & task.id %in% c("arrhythmia", : object 'FFTrees.mlr.df' not found
## Error in int_abline(a = a, b = b, h = h, v = v, untf = untf, ...): plot.new has not been called yet
ShinyFFTrees
- A point-and click (no programming), web-based version of FFTrees with around 90% of the funcionality of FFTrees
- Demo: http://econpsychbasel.shinyapps.io/ShinyFFTrees/
Publication and Collaborators
FFTrees: A toolbox to create, visualize and evaluate fast-and-frugal decision trees. (2017). Judgment and Decision Making, 12(4), 344-368.



install.packages("FFTrees") # Install FFTrees from CRAN
library("FFTrees") # Load FFTrees
FFTrees_guide() # Open the main package guide
- Manuscript: FFTrees: A toolbox to create, visualize and evaluate fast-and-frugal decision trees. Judgment and Decision Making
- This presentation: https://ndphillips.github.io/spudm2017haifa/
- Github Repository: http://www.github.com/ndphillips/FFTrees
- ShinyFFTrees: http://econpsychbasel.shinyapps.io/ShinyFFTrees/
- Email: Nathaniel.D.Phillips.is@gmail.com
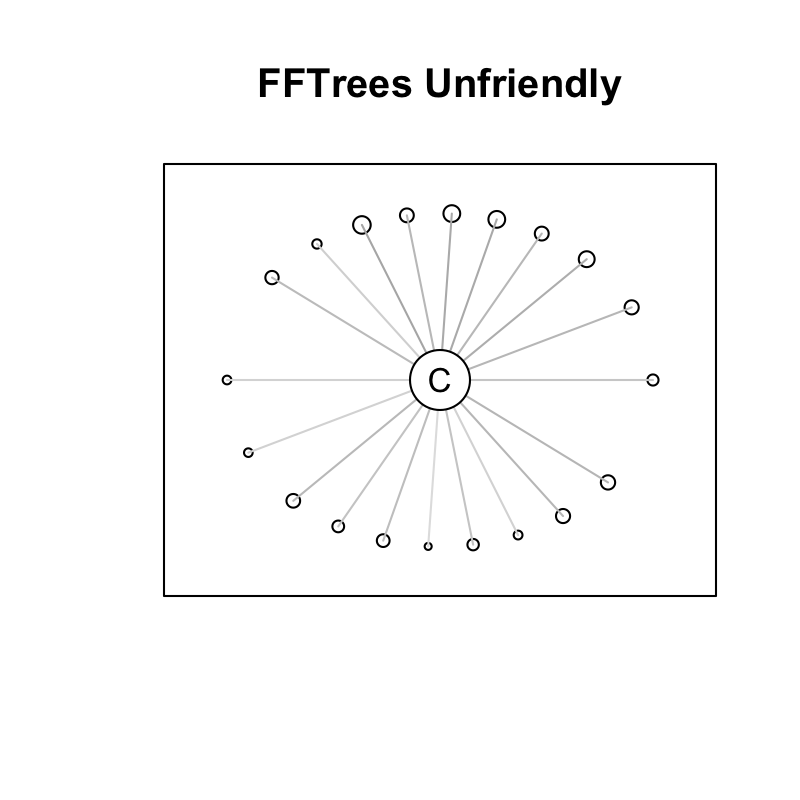
FFTrees Unfriendly Data
- Many cues, weak validity, ind errors
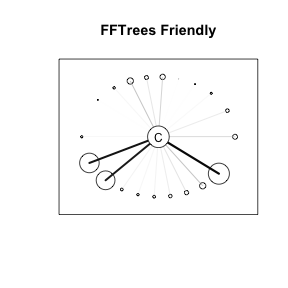
FFTrees Friendly Data
- Few cues with high validity, dep errors.
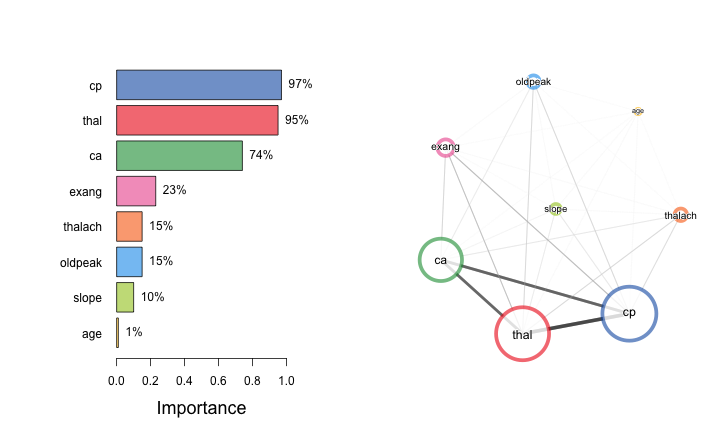
Cue importance
- As calculated by
randomForest
## Error in eval(expr, envir, enclos): object 'heart.fft' not found
## Error in rownames(heart.fft$comp$rf$model$importance): object 'heart.fft' not found
## Error in eval(expr, envir, enclos): object 'heart.importance' not found
## Error in yarrr::pirateplot(formula = importance ~ cue, data = heart.importance, : object 'heart.importance' not found
Tree Building Algorithm
- For each cue (aka, feature), calculate a threshold that maximizes
goal.chase(default: balanced accuracy) - Rank order cues by
goal.chase - Select the top
max.levels(default: 4) - Create a "fan" of all possible trees with all possible exit directions.
- Select the tree that maximizes
goal(default: balanced accuracy)
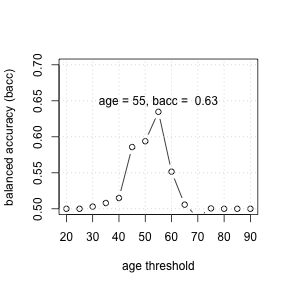
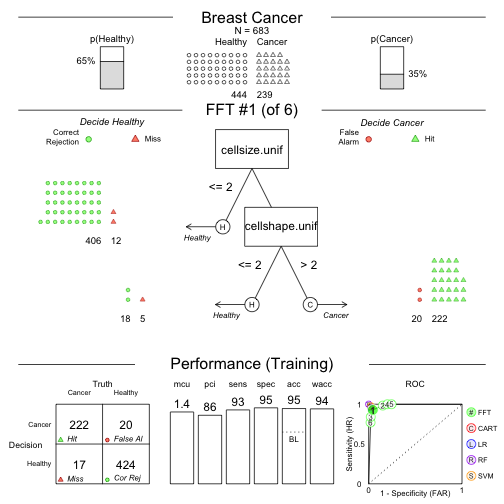
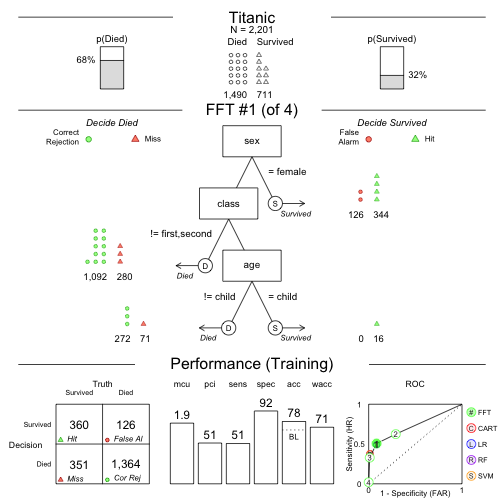
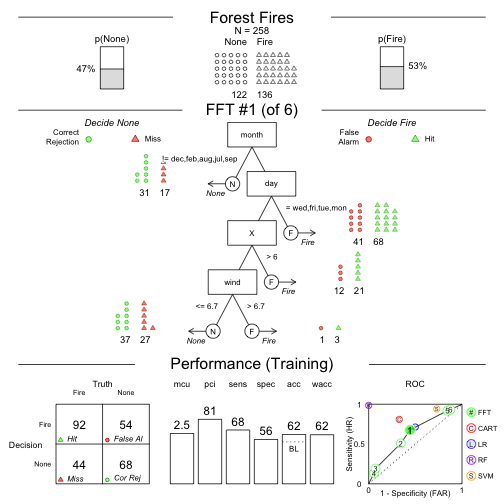
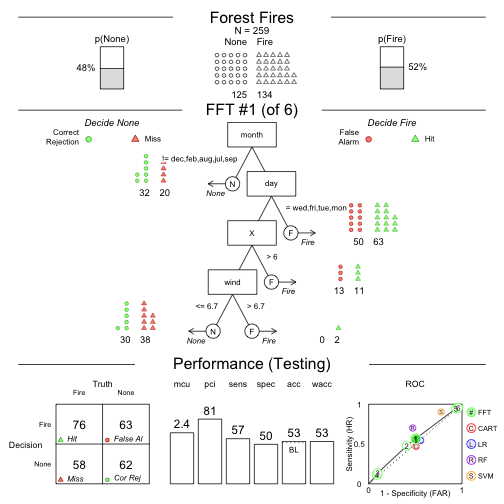
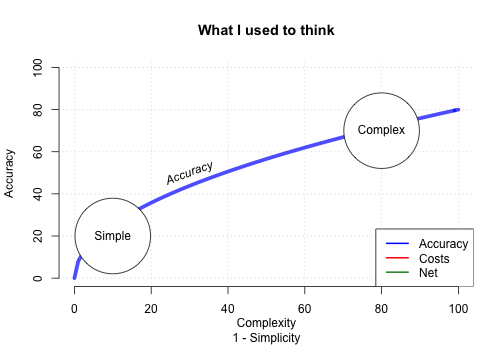
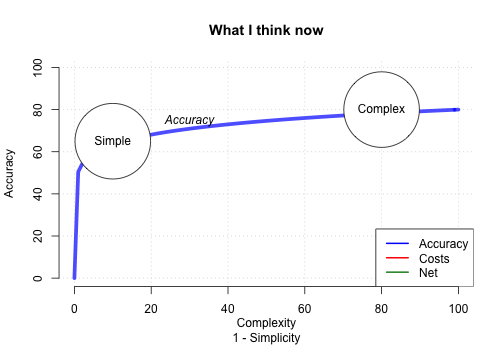
How accurate can FFTs be?
How accurate can FFTs be?
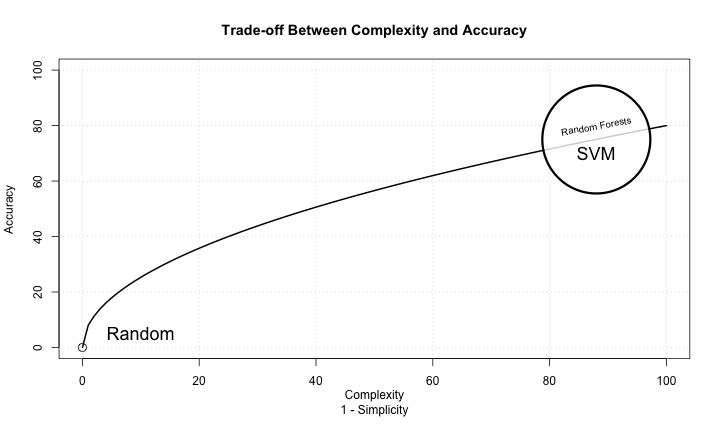
Conclusion
Why use FFTrees?
- See how, and how well, a crazy simple, transparent fast-and-frugal tree can make sense of your data.
- You might be surprised by how well it works, and generate new insights.
library("FFTrees")
a
/ \
0 b
/ \
0 1
FFTrees v1.3.2
## Growing FFTs with ifan
## Fitting non-FFTrees algorithms for comparison (you can turn this off with comp = FALSE) ...

Create a forest of FFTs
heart.fff <- FFForest(formula = diagnosis ~., data = heartdisease)